HIV genome: genetic structure and function of HIV explained
This information on the HIV genome is reproduced with permission from Molecules of HIV website by Dan Stowells. This an excellent non-technical website on explaining scientific aspects HIV and immunology. We reproduce it here to ensure it remains an online resource, but encourage people to visit the original website. All hyperlinks are to the original site.
HIV genome
The full HIV genome is encoded on one long strand of RNA. (In a free virus particle, there are actually two separate strands of RNA, but they’re exactly the same!)
This is the form it has when it is a free virus particle. When the virus is integrated into the host’s DNA genome (as a provirus) then its information too is encoded in DNA.
The following image shows roughly how the genes are laid out in HIV (remember that HIV-1 and HIV-2 are quite different).
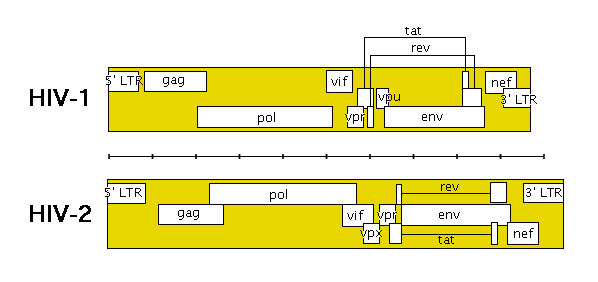
The genes in HIV’s genome are as follows:
- gag (coding for the viral capsid proteins)
- pol (notably, coding for reverse transcriptase)
(NB. gag and pol together can be expressed in one long strand called gag-pol)
- env (coding for HIV’s envelope-associated proteins)
And the regulatory genes:
- tat
- rev
- nef
- vif
- vpr
- vpu (N.B. not present in HIV-2)
- vpx (N.B. not present in HIV-1)
The HIV genome also has a “Long Terminal Repeat” (LTR) at each end of its genome – not quite a gene, but a sequence of RNA/DNA which is the same at either end and which serves some structural and regulatory purposes.
gag
gag is one of the three “main” genes found in all retroviruses (along with env and pol). It contains around 1500 nucleotides, and encodes four separate proteins which form the building blocks for the viral core:
- Capsid protein, CA, p24
- Matrix protein, MA, p17 (this protein isn’t actually part of the viral core but the “matrix” which anchors the core to the viral envelope)
- Nucleocapsid protein, NC, p9
- p6
The most significant role of the gag gene is therefore to encode important proteins which will make up the viral core.
pol
pol is one of the main retroviral genes. It encodes four proteins, of which the most important is Reverse Transcriptase. Reverse Transcriptase performs a job which is unique to retroviruses, in that it copies the virus’ RNA genome into DNA. (Since most organisms and viruses keep their genes in DNA form in the first place, they have no need to perform this task.) The copying of the HIV genome into DNA form is one of the key stages of the HIV life-cycle. The other three products of pol are these:
- Protease – which processes proteins made from HIV’s genome so that they can become part of new fully-functioning HIV particles
- RNAse H – which breaks down the retroviral genome following infection of a cell
- Integrase – which integrates the DNA copy of HIV’s genome into the host DNA
env
The env gene in HIV encodes a single protein, gp160. (When gp160 is synthesised in the cell, cellular enzymes add complex carbohydrates and turn it from a protein into a glycoprotein – hence the name “gp160” rather than “p160”.)
gp160 travels to the cell surface, where cellular enzymes again attack it, this time chopping into two pieces – gp120, and gp41. If and when new virus particles bud off from the host cell, these two pieces lie on opposite sides of the virus membrane. gp120 sits on the outside of the virus particle, forming the virus’s spikes, while gp41 sits just on the inside of the membrane – each gp41 being anchored to a gp120 through the membrane.
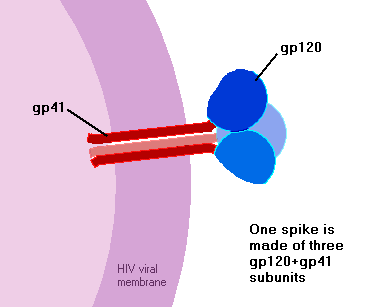
How many spikes does a HIV particle have? It’s a tricky question, but the answer seems likely to be about 9 or 10. This is a lot fewer spikes than you’ll see on most diagrams of HIV! There’s a bit of confusion since some studies have decided that HIV particles normally have 72 spikes, whilst some other studies have decided that they have normally no more than ten. It’s hard to say for certain who’s right….
tat
tat is short for transactivator – it’s a regulatory gene which accelerates the production of more HIV virus. In fact, it’s crucial to HIV, because HIV completely fails to replicate itself without it. Tat protein is also toxic, so the large amounts of tat protein released into the blood by HIV-infected cells are no help for the body.
tat works because the protein encoded by tat binds to the start of a new HIV RNA strand – a part which has been called the transactivator active region or TAR. The TAR runs from +1 to +59, that is to say, the first 59 nucleotides of the HIV genome. Once the cellular machinery has transcribed this much provirus into RNA, tat can bind to it and encourage the transcription of the remainder of the HIV genetic code.
You might have also read about the negative regulators which HIV has – NRE, nef, vif. Surely it’s barmy to have genes for boosting virus reproduction as well as genes for suppressing it! Well actually this is the normal way of things down at the level of genes and proteins. The tug-of-war between the suppressors and the activators can result in an incredibly precise control of how much a gene is expressed. Without this tug-of-war control, gene expression would simply depend on how active the cell’s transcription machinery was – either all the genes would be expressed a lot, or all the genes wouldn’t be expressed much at all. Organisms such as cells need more precision than that!
tat protein size: 101 kD in naturally-occurring HIV-1 (86 kD in some laboratory-bred types of HIV-1)
rev
rev is another of HIV’s regulator genes. It stimulates the production of HIV proteins, but suppresses the expression of HIV’s regulatory genes.
How does it achieve this? The messenger RNAs of HIV can either be sent to the protein-producing part of the cell intact, or they can have bits cut out of them first (splicing). The intact mRNA tends to encode HIV proteins (such as envelope and capsid proteins), while the spliced mRNA encodes regulatory genes such as tat and nef.
So what rev does is to help intact mRNA to be exported from the cell nucleus. It binds to the mRNA at a specific point (the RRE or rev-responsive element), and this complex of RNA and rev is sent out of the nucleus. A molecule of rev can “shuttle” in and out of the nucleus, potentially taking a new set of RNA out each time it leaves the nucleus.
The RRE is not present in completely-spliced HIV mRNA – it will have been chopped out. Completely-spliced mRNA is sent out of the nucleus by the ordinary cell machinery (without needing help from rev) – so you could say that rev’s trick is to cause the mRNA to be exported “before it’s ready”, in a sense.
nef
The negative replication factor (nef) gene encodes a protein which hangs around in the cytoplasm of the cell, and retards HIV replication. Possibly it does this by modifying cellular proteins that regulate the initiation of transcription – that is, it affects the proteins which tell the cell whether or not to make RNA copies of the DNA code.
nef protein size: 27 kD
vif
The vif gene codes for virion infectivity factor, a protein that increases the infectivity of the HIV particle.
The protein is found inside HIV-infected cells, and it works by interfering with one of the immune system’s defences – a cellular protein called APOBEC3G. Basically what happens is that vif sticks to APOBEC3G and encourages the cell to degrade it, preventing it doing its job of sneaking into newly-formed virus particles and making them non-productive.
This has been verified in experiments. If you can create a HIV virus with the vif protein missing (we would call this a delta-vif strain of HIV), then it can still infect a cell – but the new virus particles produced from that cell contain APOBEC3G and therefore aren’t very effective at infecting other cells.
vif protein size: 23 kD
Journal articles about vif:
- Navarro F, Landau NR (2004)Recent insights into HIV-1 Vif.Current Opinion in Immunology 16 (4): 477-482
- Rose KM, Marin M, Kozak SL, et al. (2004)The viral infectivity factor (Vif) of HIV-1 unveiled.Trends in Molecular Medicine 10 (6): 291-297
- Argyris EG, Pomerantz RJ (2004)HIV-1 Vif versus APOBEC3G: newly appreciated warriors in the ancient battle between virus and host.Trends in Microbiology 12 (4): 145-148
vpr
Viral protein R accelerates the production of HIV proteins.
It also facilitates the nuclear localisation of the preintegration complex – the agglomeration of viral RNA and reverse transcriptase and integrase proteins which must form in order for the HIV genome to be integrated into the host cell’s genome. vpr carries “nuclear localisation signals” (sequences of protein which are recognised by cellular machinery as indicating that it should be transported into the nucleus), and in a sense it mimicks the behaviour of a protein called importin-beta.
There also seems to be a role for vpr in stopping the host cell going through the ordinary “cell cycle” – many cells normally go through a regular cycle of splitting to create new cells, but vpr can stop host cells doing this. It seems that a cell which has been stopped during the so-called “G2” phase of the cell cycle is a nicer environment for HIV replication.
More information:
- vpr protein size: 15 kD
- There are 100 copies of this protein in every HIV virion.
- The cellular protein cyclophilin A is important for the production of vpr.
vpu
Viral protein U helps with the assembly of new virus particles, and helps them to bud from the host cell. It’s possible for HIV to replicate and bud without this particular protein, but only 10% or 20% as many new virus particles are produced.
vpu also works within the infected cell to enhace the degradation of CD4 proteins. This has the effect of reducing the amount of CD4 sticking out of the infected cell, therefore reducing the likelihood of superinfection.
Without the vpu gene, HIV virus actually kills its host cell quicker! A secondary effect of vpu is to delay the cytopathic (cell-killing) effects of virus infection, keeping the cell alive slightly longer so that it can produce more virus particles.
More information can be found in these journal articles:
- The HIV-1 Vpu protein: a multifunctional enhancer of viral particle release Bour S, Strebel K, Microbes and infection 5 (11): 1029-1039
- Functional Role of Human Immunodeficiency Virus Type 1 vpu, Ernest F. Terwilliger; Eric A. Cohen; Yichen Lu; Joseph G. Sodroski; William A. Haseltine; Proceedings of the National Academy of Sciences of the United States of America, Vol. 86, No. 13. (Jul. 1, 1989), pp. 5163-5167.
vpx
vpx is found in HIV-2 (and SIV), but not in HIV-1. It is closely related to vpr (if we compare their genetic sequences), which indicates that its existence might have come about as a duplication of the vpr gene.
Its role in the life of HIV is not entirely clear! It certainly seems to be “dispensable”, since types of HIV-2 without a functioning vpx gene still seem to be able to replicate and to infect cells…. However, it seems that vpx does have some effect of making viral reproduction more efficient, especially in non-dividing cells such as macrophages. The molecular mechanisms behind this are not yet fully understood.
More detailed information can be found in these journal articles:
- Dispensable role of the Human-Immunodeficiency-Virus Type-2 Vpx protein in viral replication, Marcon L, Michaels F, Hattori N, Fargnoli K, Gallo RC, Franchini G. Journal of Virology 65 (7): 3938-3942 JUL 1991
- Vpx and Vpr proteins of HIV-2 up-regulate the viral infectivity by a distinct mechanism in lymphocytic cells, Ueno F, Shiota H, Miyaura M, Yoshida A, Sakurai A, Tatsuki J, Koyama AH, Akari H, Adachi A, Fujita M. Microbes and Infection 5 (5): 387-395 APR 2003
Long Terminal Repeat
The Long Terminal Repeat is something which is often found in strands of RNA or DNA is the Long Terminal Repeat. At each end of the string is the same sequence of code at each end of the string. Almost like the repeat at the start and finish of these sentences, almost like!
There are two important functions for the LTR:
- Firstly they are “sticky ends” (that’s a biochemistry term) which the integrase protein uses to insert the HIV genome into host DNA.
- Secondly, they act as promoter/enhancers – when integrated into the host genome, they influence the cell machinery which transcribes DNA, to alter the amount of transcription which occurs. Protein binding sites in the LTR are involved with RNA initiation.
Last updated: 2 August 2010.

