9. Appendix 6: Stanford Drug Resistance Database online tables
The Stanford Drug Resistance Database is one of several online research resources that contain a vast amount of technical information about drug resistance.
It includes links to each drug family (drug resistance mutation tables) and the related mutations and individual pages for each HIV drug (antiretroviral drug summaries).
The tables are summarised as a single page. (October 2022).
Notes and comments on mutations are organised by drug class.
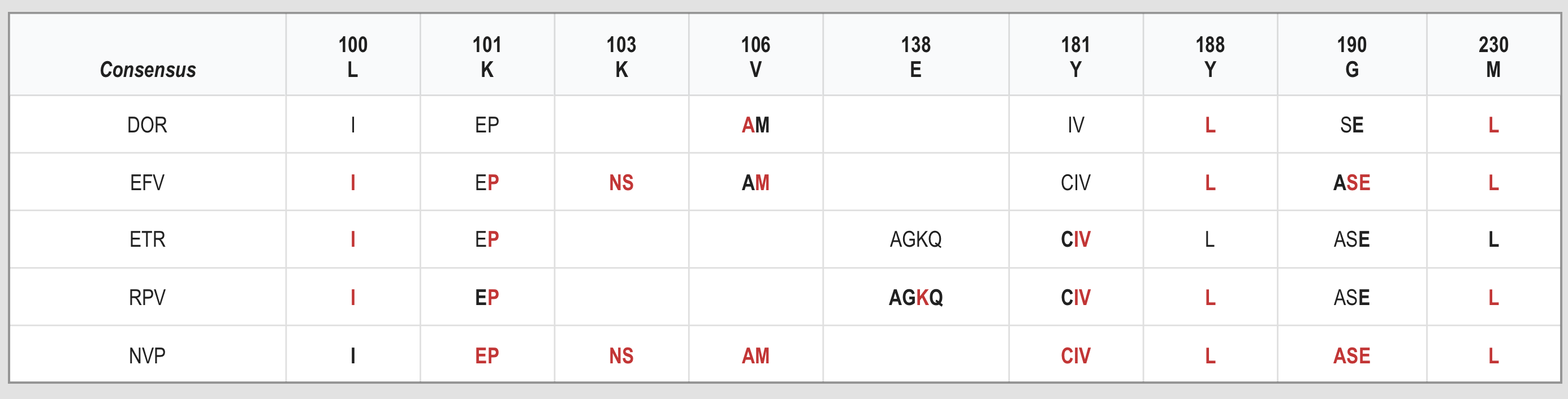
These tables are complicated but they are explained in notes for each mutation. (See Table 4).
Table 4: Integrase inhibitor (INSTI) resistance mutations (Stanford database, 2022)
The consensus sequence means wild-type non-resistant virus. The letters in the Consensus row refer to the amino acids that should be at those junctions (codons).
The line of numbers above the ‘Cons’ row are the positions or junctions where important changes occur.
Each of these junctions (called a codon) in integrase is linked to resistance to different integrase inhibitors.
The letter in the main body of the table shows the amino acid changes at each codon, Letters in bold show that this is an important mutation in reducing how well that drug will work. Letters in red indicate high levels of resistance that stop the drug from working at all.
Each row is for a different integrase inhibitor. If codon 118 changes from G to R (ie mutation G118R), then cabotegravir, raltegravir and elvitegravir will no longer work and dolutegravir and bictegravir will be less effective.
The detailed comments on G118R below include more detail about this mutation mainly developing in people who are on dolutegravir, and on levels of resistance for each INSTI.
Last updated: 1 January 2023.