9.4 Resistance tests and interpreting test results
9.4.1 Recap of previous section
The previous section looked at how HIV can evolve differently depending on whether or not you are on ART.
If viral load continues to be detectable on ART this leads to drug resistance.
Also, that HIV has a very rapid and prolific life cycle.
Instead of thinking about having one virus, we talked about how someone really has thousands of slightly different viruses. The dominant virus will depend on the HIV drugs that someone is taking. Starting, continuing or stopping ART changes the environment and this can change the dominant virus.
When drug levels are too low to keep viral load suppressed, HIV can develop resistance to one of more of the drugs in ART. The resistant virus is now relatively more fit than wild-type HIV and so it becomes the dominant or majority.
If viral load continues to increase on failing ART, more complicated patterns of mutations can develop because of the ‘selective pressure’ of the drugs. Combinations of mutations evolve in ways that would be unlikely if drugs were not present.
On effective ART, new resistance only develops when viral load is detectable (greater than 50 copies/mL). If viral load stays undetectable and adherence is good, then resistance will be rare.
Learning how resistance tests work will help you to understand this.
9.4.2 Introduction to section 4
Section 4 explains different types of resistance tests and how to interpret the results.
The main type of test is called a genotype test. Phenotype and virtual phenotype tests are now rarely used in the UK, other than in research.
Interpreting the results of these tests can be difficult and complicated. We include an outline and why experts are needed to do this.
Both genotype tests and phenotype tests work from blood samples but they work in different ways (see Figure 5). The results are also interpreted differently.
Figure 5: How genotype and phenotype tests work
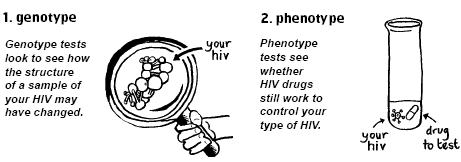
These two words are often used in other aspects of science.
Genotype refers to the genetic sequence and specific changes. Phenotype refers to observations related to the changes (i.e. in a test tube).
Section 4 also includes technical information, so might take longer. Please write down questions if anything is not clear.
9.4.3 Genotype tests and genotypic resistance: numbers and letters
Genotype tests (also called genotypic tests) look for changes in the structure of HIV.
The test compares genetic sequences to wild-type HIV. Mutations are described with numbers and letters.
This is easier to imagine if you think of HIV as a long chain of letters that were talked about in Section 2. Each letter (A, C, T or G) represents a base that for HIV in just over 9000 letter long.
Each group of three bases is an amino acid. The example in Appendix 2 shows that the three bases for the amino acid called methionine (M) only needs one base change to change the amino acid to valine (V). This mutation is a simple change.
If the three bases change from ATA to GTA, the amino acid at that junction changes from methionine to valine. Appendix 5 illustrates mutations in relation to the HIV genome.
The section of the genome used by nukes (and NNRTIs) is called reverse transcriptase (RT). If the change for ATA to GTA occurs at junction number 184 in the RT gene, this will affect how some drugs work.
The mutation described above is written as M184V in RT.
If the virus changes at junction 103 on the RT genome, from AAA or AAG to AAC or AAT, the amino acid at that junction changes from lysine (K) to asparagine (N). This mutation is written as K103N (in RT).
You don’t need to learn these names but a key to the abbreviation letters for different amino acids is in Appendix 3.
These two mutations are good examples to start with.
- They only need one base change to change the amino acid.
- They are both very common mutations
- Their impact is like an on-off switch. Without the mutation the drug works. With the mutation the drug doesn’t.
M184V results in high level resistance to 3TC and FTC. It stops both these drugs from working.
K103N results in high level resistance to NNRTIs like nevirapine and efavirenz. It stops both these drugs from working.
Genotype tests work by comparing the genetic code of a sample of HIV to the code that would be there in wild-type (WT) HIV that has no drug resistance.
Results from a genotype resistance test come as a list of the differences that are found in the sample. They should also include a note explaining the importance of each mutation.
Table 1 lists key mutations and the impact they have on treatment. The integrase mutation is included as a more complicated example.
Table 1: Important drug resistance mutations
| Mutation | Genome | Interpretation |
| M41L | RT | M41L usually occurs with T215Y. Together these mutations confer intermediate-to-high resistance to AZT and d4T and a lower resistance to ddI, abacavir, and tenofovir. |
| K65R | RT | K65R causes intermediate resistance to ddI, abacavir, 3TC, FTC, and tenofovir, and low-level resistance to d4T. K65R causes AZT to be more active (called hypersensitive or hypersusceptible). |
| M184V | RT | High level resistance to 3TC and FTC and low level resistance to abacavir and ddI |
| K103N | RT | K103N causes high-level resistance to nevirapine, delavirdine, and efavirenz. By itself it does not affect etravirine efficacy. However, it increases the effect on etravirine from 3-fold to 15-fold reduced sensitivity when L100I is also present. |
| I50L | PRO | I50L causes intermediate-to-high level resistance to atazanavir/r and increases susceptibility to other PIs. |
| L90M | PRO | L90M causes resistance to nelfinavir, saquinavir/r, atazanavir/r, and indinavir/r. When present with other mutations it also reduces the activity of fosamprenavir/r and lopinavir/r (Kaletra). |
| G118R | Integrase | G118R generally only develops while taking dolutegravir. It prevents cabotegravir, elvitegraivr or raltegravir from working (high level resistance) and reduces the effectiveness of dolutegravir and bictegravir (partial resistance). |
| Q148H/K/R | Integrase | Q148H/K or R are mutations selected by raltegravir and elvitegravir. By itself Q148H reduces susceptibility to both these drugs by about 5-10 fold and Q148RK reduces susceptibility by >30-100 fold. With G140S, Q148HRK reduces susceptibility by more than 100-fold. Q148HKR alone have minimal effects on GSK572 but causes more than 10-fold reduced susceptibility in combination with E138K, with or without G140S. |
Key point 5: Some mutations stop a drug from working completely (high level resistance). However, some only have a moderate impact (intermediate) and some only have little impact (low level resistance).
Key point 6: Mutations that are associated with resistance to one drug can also have resistance to similar drugs in the same family. This is called ‘cross-resistance’. For example, if you develop resistance to one NNRTI like efavirenz it is likely you will be cross-resistant to nevirapine, even though you have never taken nevirapine.
There are too many mutations to remember but it is good to know the common examples in Table 1.
Luckily, several research groups publish comprehensive tables and explanations online. Use these if you need to find about about a specific mutation or drug. See: Appendix 6: Stanford Drug Resistance Database online tables.
As you learn more about drug resistance, it gets easier to remember key mutations, especially knowing what the letters and numbers mean.
The IAS-USA guidelines illustrate resistance to each drug in a different way.
![]()
Each drug has a bar representing the section of the HIV genome where resistance develops. The numbers inside the bar are the junctions where mutations are linked to resistance. The letters on the top are the amino acids at that junction for wild-type HIV. The letters underneath are the amino acid changes linked to drug resistance.
This is a good way to visually compare the resistance profile of drugs in the same class. It is a quick way to get an idea of cross-resistance. Appendix 7 includes these charts and links to the original resources.
Mutations that show resistance is reverting back to wild-type are called revertant mutations.
For example, the T215Y mutation is associated with AZT resistance. If this mutation is transmitted to someone who is not taking AZT, the virus changes back closer to wild type. The mutations T215D, T215N or T215S are seen as ‘tracks’ in this change. They are interpreted as previously having T215Y and imply resistance to AZT.
Question: How do researchers find out about the mutations associated with each drug?
Answer: Every new drug is tested in test tube studies to see which mutations occur (called in vitro passaging). These are usually similar to the mutations seen in human studies when treatment fails (ie in people whose viral load stays detectable). For example, in studies that include 3TC, the M184V mutation is one of the first changes seen if viral load is not reduced to undetectable (less than 50 copies/mL). Using 3TC without any other HIV drugs (i.e. 3TC monotherapy), results in M184V within a couple of weeks. This 3TC resistant HIV will be cross-resistant to FTC. New drugs are often developed to specifically work against resistant virus.
Question: Are resistance tests perfect? Are all mutations known?
Answer: Resistance tests are not perfect, but major mutations usually accurately predict when a drug will not work. Given the number of possible combinations of mutations, this is an area of research that is always developing.
9.4.4 Phenotype tests and phenotypic resistance: x-fold resistance
Phenotype resistance tests look at resistance in a very different way.
Instead of looking at mutations, phenotype tests measure how active a drug is in a sample of HIV compared to how active it is in a sample of wild-type HIV.
An HIV drug is added to a sample of HIV in a test tube, and the test measures how much HIV continues to be produced. The quantity of the drug is then slowly increased to see how much extra is needed to have the same impact on reducing viral replication compared to a normal dose on wild-type HIV.
For example, the T215Y mutation is associated with AZT resistance. If this mutation is transmitted to someone who is not taking AZT, the virus changes back closer to wild-type. The mutations T215D, T215N or T215S are seen as ‘tracks’ in this change. They are interpreted as previously having T215Y and so are resistant to AZT.
Phenotype results are given as a fold change (or cut-off) with an interpretation of what this means. For example, if a sample needed four times the quantity of drug to have the same impact on stopping the virus replicating, the result would be 4-fold resistance (or 4-fold loss in sensitivity). In practice, you would need to increase the daily dose by four times to get the same effect on viral load.
Sometimes resistance can be overcome by increasing the dose of medication in a person. In practice, this only tends to be when someone has complicated resistance and fewer drug choices. For most drugs and in most circumstances this would cause too many side effects to be an option. Some HIV drugs, including darunavir and dolutegravir, have higher doses for people with drug resistance.
The clinical impact of phenotypic resistance varies depending on each drug. For example, 4-fold resistance to one drug may still be sensitive while 4-fold resistance to another may be resistant. Each drug has a different cut-off for when a drug is sensitive, partially resistant or completely resistant. Nukes generally become resistant at low fold-changes while PIs have higher thresholds.
Each make of phenotype test has its own reference chart for cut-off values for each drug. These numerical values can differ between tests.
9.4.5 How are genotype and phenotype resistance related?
The clinical interpretation of both genotype and phenotype resistance tests involves deciding on which drugs to use for any one individual.
This is supported by results collected in large databases with the results from thousands of previous resistance test results.
These databases relate the average impact that a mutation (or pattern of mutations) has on the phenotypic resistance.
For example, M184V is associated with high level resistance to 3TC and FTC, and results in a more than 300-fold reduction in drug sensitivity. With this mutation, it would take a dose 300 times higher than the standard dose to have the same anti-HIV effect. This would be physically impossible and too toxic.
M184V may still have a benefit in continuing 3TC because M184V replicates less well than wild-type and because it increases susceptibility to AZT, d4T and tenofovir. However, the benefit of continued 3TC will be less than the benefit of 3TC in patients with wild-type HIV.
This is an unusual feature of M184V that is not shared with most other mutations. Although other mutations also reduce fitness, this effect is usually overcome by new mutations that compensate for this.
Continuing to take 3TC or FTC to keep the M184V mutation may keep viral load a little lower. This is because the virus is less fit. It reproduces less well so there is less virus.
9.4.6 Virtual phenotype tests
Virtual phenotype tests are a third type of resistance test.
They are really genotype tests, and the mutations that are detected are included in the results. However, the pattern of mutations is also compared to a huge database of matched genotype and phenotype results and a phenotypic result is predicted from the database.
As with the other tests, an interpretation comes with the results that explains whether each drug is likely to be sensitive, intermediate or resistant.
These tests produced very sophisticated individual results. However, they are now rarely used because drug options to treat resistant HIV are now much better. One of the main virtual phenotype tests, produced by Virco, was withdrawn at the end of 2013 becasue of low use.
9.4.7 Primary and secondary vs major and minor mutations
You may read about primary and secondary resistance mutations, but these terms are used more rarely now.
Instead, major and minor mutations are preferred terms.
Major mutations have a big impact on drug resistance.
Minor mutations only have a small impact on drug resistance.
The terms primary and secondary are confusing because they sound like primary mutations occur first. Sometimes however, the first mutations make little difference.
For example, with protease inhibitors the first mutations have little impact on how well a drug works. Then, as more mutations accumulate, the impact becomes more important.
Finally, after 5 or 6 or more mutations, the clinical impact becomes more significant. With protease inhibitors the first mutations to occur are minor (secondary) mutations and major (primary) mutations occur later.
9.4.8 Resistance testing: practical issues
Resistance testing is widely used in most western countries. The information they provide helps choose drugs that have the best chance of working.
However, resistance tests are more accurate in showing which drugs will not work, than guaranteeing which drugs will work.
- Resistance tests can only detect resistance to drugs that you are currently taking or have recently been taking. Remember that when a drug is stopped, wild-type HIV becomes relatively more fit (see section 3) and many mutations reduce to levels that are too low to detect. Usually a mutation has to be present in more than 20% of viral population to be detected with routine tests.
- Treatment choice needs to be based on someone’s lifetime history of treatment and resistance, not just the single result of one current resistance test.
- In addition to the report given by the test lab, results need to be interpreted by an expert who has your treatment and resistance history. Experts do not always agree and different databases sometimes report different results. Even though the tests may not all agree, it is better to have this information to inform your treatment choices.
- There is less information about resistance to newer drugs. This is because fewer people have developed resistance to those treatments. There is less information in resistance databases to predict how new or old mutations will affect how these drugs will work.
9.4.9 Section 4: Learning points
This has been a complicated section.
- There are three main types of resistance tests but genotype tests are used most frequently.
- Different mutations have different clinical implications. Some are associated with high level resistance, some with intermediate and some with low resistance.
- Resistance for some drugs accumulate steadily on an increasing scale. But for other drugs, it only takes one key mutation to devlop complete resistance.
- Genotype tests report mutations and phenotype tests report fold-changes. All resistance tests should include a detailed interpretation for each drug. You can ask for a copy of this report.
- Resistance to one drug can result in resistance to similar drugs in the same class. This is called cross-resistance.
- Viral load needs to be detectable to get a result. How ‘detectable’ depends on the specific lab and test. This used to be above 500 copies/mL but some labs can get a result when viral load is between 50 and 500 copies/mL.
- The interpretation of complicated results requires expert advice.
9.4.10 Section 4 evaluation
Please now take a few minutes to evaluate this session online.
This single page includes six short questions.
Your answers are anonymous, but the course is dependent on your feedback, which is appreciated.
Last updated: 1 January 2023.
